The benefits of misbehaving: prion helps yeast survive environmental stress
Motomasa Tanaka and colleagues show that prions help yeast cells survive drug treatment, revealing a functional role for protein aggregates.
June 26, 2012
Prions are aberrantly folded proteins that catalyze the misfolding and aggregation of other proteins. In a recent report in Science, Motomasa Tanaka, Team Leader of the Laboratory for Protein Conformation Diseases at RIKEN BSI and his colleagues demonstrate that the yeast prion Mod5 confers yeast with resistance to certain drugs. This is the first report of a functional role for prions in the cell and suggests that protein aggregation can allow rapid cell adaptation to stressful environmental conditions. This finding has potential implications for understanding and treating human neurodegenerative diseases associated with misfolded protein aggregates.
Like origami, the folding of a protein from a linear string of amino acids into a complex and functional three-dimensional shape, must follow certain rules. Each fold contributes to the final structure. Misfolding can lead to an unrecognizable clump of paper in one case and a nonfunctional and possibly toxic protein in the other. Normal cells have mechanisms for identifying and degrading misfolded or otherwise damaged proteins. However, these mechanisms are not failsafe and sometimes, having escaped degradation, misfolded proteins form aggregates. Proteins sequestered in aggregates cannot perform their normal function, effectively making the cells lose that function. Therefore aggregate formation is generally regarded as detrimental to cells.
One type of aggregate, called amyloid, has a highly ordered, fibrillar structure made up of proteins rich in β-pleated sheets. Recent studies show that many proteins have the potential to assemble into amyloid. In most cases, human proteins that form amyloid, especially in the brain, are considered indicators of neurodegenerative diseases. Some examples include the alpha-synuclein protein in Parkinson’s disease, Aβ42 in Alzheimer’s disease and the huntingtin protein with expanded polyglutamine repeats in Huntington’s disease. Prion diseases — the transmissible spongiform encephalopathies (TSE), such as Creutzfeldt-Jakob disease (CJD) in humans, scrapie in sheep and bovine spongiform encephalopathy (BSE) in cattle — are also considered part of this group. The infectious prion protein (PrP) also forms amyloid aggregates, and can be transmitted between individuals and even across species.
Prions, short for “proteinaceous infectious particle,” were identified in the 1980s in humans suffering from CJD and other TSEs. Subsequently, prions were discovered in budding yeast, the study of which, owing to the availability of a large array of experimental tools, accelerated the characterization of their biochemical properties and mechanisms. However, unlike mammalian prions, most yeast prion proteins do not greatly hinder cell growth. Hints from earlier research suggested that fungal prion aggregates may have physiological functions, however this possibility had never been clearly demonstrated.
Searching for a new type of prion
One of the research goals of the Tanaka lab is to identify novel prions with physiological roles in cells. All yeast prions known prior to this work (about 10) have domains rich in the amino acids glutamine and asparagine (Gln/Asn-rich). This so-called aggregation-inducing domain can readily misfold into β-sheets and makes it easier for prions to assemble together. However, the human prion protein does not have a Gln/Asn-rich domain, which led Tanaka to speculate that non-Gln/Asn-rich prions should also exist in yeast.
“I wanted to fill the gap by identifying non-glutamine/asparagine-rich yeast prion proteins,” says Tanaka. To find these elusive prions he and his colleagues, postdoc Genjiro Suzuki and technician Naoyuki Shimazu, designed a novel screen to identify proteins able to form amyloid-like aggregates. They utilized their knowledge of another class of neurodegenerative diseases studied in the lab, collectively called polyglutamine diseases, because the aggregation domain in the proteins associated with these diseases is made up of long stretches of the amino acid glutamine. In this system, polyglutamine acted as a reporter to identify aggregate-forming proteins.
“Polyglutamine easily forms aggregates. The soluble polyglutamine protein can form aggregates using the seed aggregate of another protein as a template. I thought that I could use this cross-seeding system to detect the aggregation of yeast proteins,” says Tanaka.
Out of about 6000 genes screened, Shimazu identified 65 proteins as potential prions. Those that were known to have Gln/Asn-rich domains or other repeat sequences were excluded, along with others that were essential for life, loss of which was lethal to the cells. In the end, Mod5, which was not previously known to be a prion, was chosen for further analysis.
The confirmation that Mod5 was indeed a prion involved a battery of assays such as a demonstration of amyloid formation in vitro and prion conversion in vivo. The aggregation domain of Mod5 was novel and Suzuki was able to identify it with the help of Yumiko Ohhashi, another postdoc in the Lab, who studies protein misfolding using a biophysical approach. Yeast strains with a mutant Mod5 that lacked the aggregation domain did not efficiently form amyloid, confirming its requirement for amyloid formation.
Adapting to a stressful environment
In yeast, insoluble prion aggregates do not induce severe cytotoxicity, unlike the prion protein in humans. For a number of years, there had been speculation that aggregated prions in yeast might have a physiological role, but exactly how this state might be advantageous was not known.
“I was thinking that I would like to prove that active conversion from the normal to the prion state acquires novel cellular functions,” says Tanaka. Mod5 is a tRNA isopentenyltransferase, and adds an isopentenyl modification to the AUU anticodon of tRNA using the substrate DMAPP (dimethylallyl pyrophosphate). The same substrate is also used by the enzyme Erg20 in the sterol biosynthetic pathway.
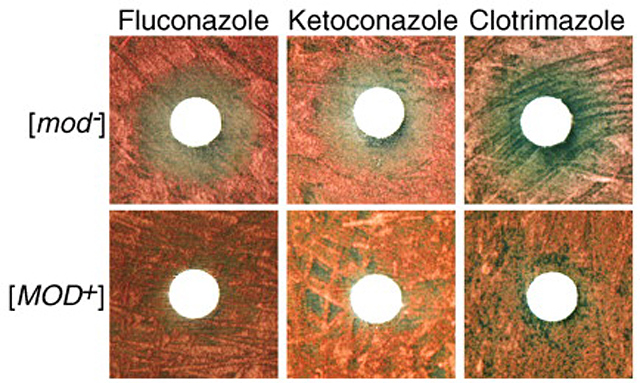
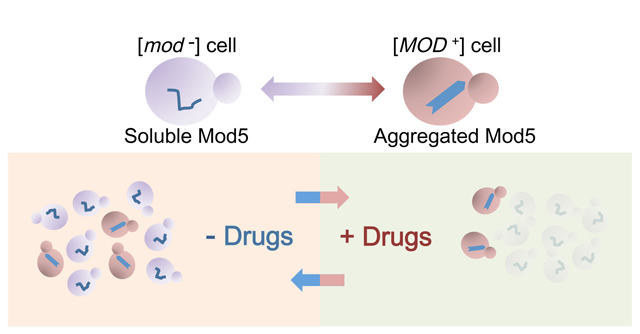
“We examined the literature and databases and found that the loss of Mod5 may increase the level of ergosterols. We tested this assumption in yeast and found that cells with Mod5 in the prion state [MOD+] indeed have higher ergosterol levels. This means that the [MOD+] yeast cells have thick cell membranes to protect them from something. The yeast might have acquired resistance to some drugs.” A search among known drugs revealed that [MOD+] yeast were resistant to a class of antifungal drugs that inhibit ergosterol synthesis. Essentially, the aggregation of Mod5 into an insoluble amyloid acts as a switch between the tRNA modification and sterol biosynthesis pathways, shutting down the former and enhancing the later.
The [MOD+] state confers a selective advantage to yeast so that in a population of [mod–] yeast grown on antifungal drugs [MOD+] yeast cells arose at a higher rate than they did otherwise. In fact, in the presence of drugs prion conversion was enhanced, suggesting that prion conversion may be utilized by yeast in times of stress to quickly adapt to changes in the environment without the more lasting and deleterious effects of DNA mutation.
Tanaka’s discovery that prion conversion can make fungi resistant to drugs could lead to novel antifungal treatments for pathogenic fungi by preventing prion conversion. However, it remains to be demonstrated whether other prions can provide a similar drug-resistance benefit and if the same is true of other fungi. Nonetheless, these findings raise the possibility of harnessing the benefits of prion conversion for applications in medicine and agriculture.
Focusing on human brain diseases
What parallels, if any, can be drawn between yeast prions and mammalian prion disease and neurodegenerative disorders? Tanaka’s research indicates that yeast can use prion aggregation as an adaptation for responding to environmental stress. Could amyloid formation in human cells also represent an adaptive response rather than the cause of disease? In recent years, researchers have begun exploring the possibility that amyloid itself is not responsible for disease but is merely an end product of the aggregation of toxic oligomers or monomers.
“It remains controversial whether amyloid is toxic or non-toxic,” says Tanaka. Recently many papers suggest that in Alzheimer’s disease or other neurodegenerative diseases the toxic species may be protein oligomers or monomers rather than amyloid.” He adds: “The implication of our work for neurodegenerative diseases is that prion conversion may be a stress response. In mammals, the neuron may try to cope with stress by converting more toxic soluble oligomers or monomers into less toxic amyloid forms.”
Although the search for other functional amyloids in yeast continues to be a focus of the Tanaka lab, he is also interested in the structural details of prion conversion from a soluble monomer to an insoluble amyloid state, because this structural conversion is common to other neurodegenerative disorders but not much is known about the underlying mechanism.
“I want to understand how protein misfolding occurs and how these structural defects result in protein aggregation and disease,” says Tanaka. “I try to challenge these fundamental questions in biology. Fortunately, the postdocs in my lab have different backgrounds and this allows us to apply diverse approaches in various model systems.”
When asked about his motivations as a scientist in the amyloid field he says: “The reason why I study protein misfolding and aggregation is because I want to reveal the mechanisms of neurodegenerative and psychiatric diseases.” He adds, “I would be very glad if our research could contribute to society.”
By Alexandra Terashima
© RIKEN BSI 2012
Image credits (Photo: A. Terashima; Figures: M. Tanaka)
To learn more about research opportunities in the Tanaka lab, please visit their website.
Read the article:
Genjiro Suzuki, Naoyuki Shimazu and Motomasa Tanaka. (2012) “A Yeast Prion, Mod5, Promotes Acquired Drug Resistance and Cell Survival Under Environmental Stress.” Science, 336 (6079): 355-359. DOI: 10.1126/science.1219491
For additional information about the Tanaka lab:
![]() Motomasa Tanaka Lab Website
Motomasa Tanaka Lab Website
![]() RIKEN Press Release
RIKEN Press Release
![]() RIKEN Research Interview with Dr. Tanaka
RIKEN Research Interview with Dr. Tanaka
For more information about prions:
R. B. Wickner, H. K. Edskes, F. Shewmaker, T. Nakayashiki. (2007). “Prions of fungi: inherited structures and biological roles.” Nat. Rev. Microbiol. 5 (8): 611-618. PMID: 17632572
M. F. Tuite, T. R. Serio. (2010) “The prion hypothesis: from biological anomaly to basic regulatory mechanism.” Nat. Rev. Mol. Cell Biol. 11 (12): 823-22. PMID: 21081963
R. Halfmann, S. Lindquist. (2010) “Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits.” Science, 330 (6004), 629-632. PMID: 21030648
MDavid W. Colby and Stanley B. Prusiner. (2011) “Prions.” Cold Spring Harb Perspect Biol. DOI: 10.1101/cshperspect.a006833
Staniforth GL and Tuite MF. (2012) “Fungal prions.” Prog Mol Biol Transl Sci. 107, 417-56. PMID: 22482457