In the past five years, researches have sequenced the genomes of several different animal species, including humans. While some of these sequences are not complete, these advances have helped us uncover many genes as well as transcribes that do not code for proteins. Each chromosome is a giant DNA molecule that contains millions of nucleic acids. Our lab's goal is to find the regulatory mechanisms of genes on the chromosome.
Most cells in an organism (excluding those in the immune system) contain the same chromosomes. However, gene expression in each cell varies. When a gene is needed, its code is read and extracted from the chromosome and the gene is called into action: the latter is commonly called gene expression. For example, specific genes are expressed in neural cells during fetal development that are not expressed in healthy adult brains, and visa versa. In fact, many developmental anomalies and diseases are probably triggered by inappropriate gene expression. The proper reading of a gene at the right time is essential to avoid such disorders.
According to current understanding, the DNA sequence (or domain) that defines the start of a specific gene's expression regulates the genetic expression (or transcription) of that gene within a given cell. This domain is called the promoter and the domain that turns the promoter on or off is called the enhancer. Since a chromosome is a giant DNA that contains large numbers of genes, the domains within can be either promoters or enhancers. Yet, we do not know how a coupled promoter and enhancer might interact. Recent research identified regulatory elements that exist far apart-as far as one megabases from a promoter. It is not known how these two regulatory sequences-promoter and regulatory element interact to achieve proper expression of a gene.
Cell proliferation occurs rapidly in development and requires a large number of genes to be expressed at specific times as the organism grows. Therefore, genes must be switched on and off dynamically. Errors in this switching produce anomalies in tissue development or in function. We want to clarify the regulatory mechanism within the chromosome. To do this, we have targeted Hox genes, which determine identity the body structure along anterior-posterior axis.
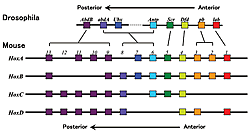
For mammals, 39 genes are present in four chromosomal domains, forming clusters of about 10 genes.
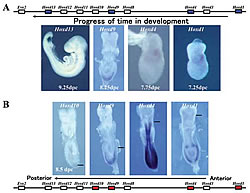
A: Start of expression of Hoxd1, 4, 9, 13 according to the time in development (genes marked blue). The start of gene expression corresponds to the order of genes on the chromosome.
B: Expression of Hoxd1, 4, 9, 13 with a mouse fetus 8 days after fertilization (genes marked red). A line is drawn on the forefront of each gene-expressed domain. At this time, Evx2 is not yet expressed from Hoxd11
Genetic studies on the fruit fly showed that Hox genes play an important role in body formation. Eight genes have been identified for the Drosophila, while mammals have 39 known genes (Fig.1). All of these genes are transcription regulatory factors with a DNA binding domain called the homeobox. These genes are clustered in a certain part of the chromosome. In mammals, there are groups (or complex) of about ten genes that are clustered at the same transcription direction of four chromosomal domains with lengths of about 100 kb (Fig.1). The order of the gene sequence along the chromosome reflects the order of gene expression. At one end, a gene will be expressed in the earliest stages of development (about 7.0 days after fertilization in mice), while the other genes are expressed in sequence according to their positions in the chromosome until the last gene is expressed (at about 9.5 days after fertilization in mice) (Fig.2). Similarly, this gene expression pattern also reflects the chromosomal structure (Fig.2). Therefore, at least for the Hox genes, a strict selection process governs enhancer-promoter interaction within this dense concentration of genes. Furthermore, researchers have found an enhancer that is more than 300kb from the Hox gene complex. Concerted effort from various labs has exposed the relationship between the position of the gene on the chromosome and the regulation of its expression. Now, we are trying to determine how regulation timing is mainly dependent on a mechanism that regulates the entire complex. The rules governing the interaction between the enhancer and promoter are also being clarified. Nevertheless, the complexity of these processes have exposed inconsistencies in our understanding-revealing just how much research still needs to be done.