Professor Okazaki, one of the collaborators in this study, and his colleagues, discovered that genomes are different by investigating the discordant occurrence of mental illness in monozygotic twins in 1998. Inspired by this novel approach, I sought to determine the causes bipolar disorder by studying identical twins that are discordant for this mental disorder. This proved difficult, however, because such occurrences are rare and no method to identify minute differences between genomes had yet been established. A fellow researcher, Dr. Kakiuchi, proposed a twin study shortly after I was appointed as a laboratory head at RIKEN BSI. The timing and facilities at this Institute were ideal for the kind of study I had wanted to undertake to determine genomic differences between twins using then new technology: DNA microarray. With this technology it was possible to investigate tens of thousands of genes simultaneously.
Now, it was simply of matter of collecting samples from discordant twins. I began asking acquaintances at every meeting I attended if they were working with twins with bipolar disorder; going so far at to ask, after delivering a speech at a an alumni reunion, attendees about their twin patients. As bipolar disorder typically develops in both twin siblings, discordant development of the disorder is uncommon and valuable. Fortunately, a doctor attending a small meeting told me that he was currently attending a patient whose twin was not affected with the disorder, and finally our project could begin.
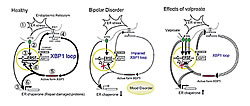
Middle: A difference in XBP1 binding sequence, where C becomes G, inhibits XBP1 binding and prevents XBP1 loop function. Inhibiting the XBP1 loop might cause mood disorders.
Right: Valproate, a mood stabilizer, compensates for XBP1 loop dysfunction by increasing ATF6 which activates the XBP1 loop.
ERSE: endoplasmic reticulum stress element
Dr. Kakiuchi, who arrived in June 2001, first incubated cells extracted from the blood of two pairs of identical twins that were discordant for bipolar disorder and examined their gene expressions using DNA microarray. He identified a pair of genes associated with the endoplasmic reticulum stress pathway: XBP1, located at 22q12 which is strongly linked with bipolar disorder; and GRP78 whose expression is regulated by XBP1. As levels of GRP78 are reportedly elevated in response to bipolar disorder treatments, our findings seemed sound.The endoplasmic reticulum stress pathway is the process by which an overload in damaged proteins is repaired. By inducing ER stress in cells, we observed differences in the responses of cells from bipolar disorder patients and those from controls. Investigating the causes of these differences, we found that individual differences in the upstream promoter sequence for XBP1 expression variably triggered a feedback loop, the XBP1 loop, to produce self-binding XBP1 protein and GRP78 that repair damaged proteins.
If the resulting depression to ER stress contributes to the development of bipolar disorder, what affect will mood stabilizers have on this response? In cells cultured with mood stabilizers, only valproate improved the ER stress response that had been derailed by the polymorphic gene. If these results are verified in labs around the world leading to clinical studies, we might be able to develop a system that will predict the effects of mood stabilizers in treating this disorder. Furthermore, if the action mechanisms of different mood stabilizers can be identified, it will be possible to develop new medications for treatment.
In my 14 years of research on bipolar disorder, this was the first result I had that had direct implications for treating patients. The response from researchers not directly connected to mental illness research has also been surprising. Should our findings directly contribute to alleviating the burden of mental illness, the real benefit of our investigations will be realized.