How does the brain develop? Is there a master plan guiding its development? These are questions that seem to be interest to anyone. Some researchers have recently starting investigating the human genome in the pursuit of answers. The genome encodes the genetic instructions of life. Since the latter half of the 1990’s, the genomic sequences of various organisms have been determined one by one, and, in October 2004, researchers announced that the euchromatic sequence of the human genome was nearly complete. (Euchromatin refers to the gene rich portion of the genome that was incomplete in the original draft sequence.) This historic achievement also heralded a revision to the estimated number of protein-encoding genes to around 22,000. The genome also contains information on the basic design of the brain, including cellular differentiation, formation of the basic circuitry, and regionalization which is regulated by specific genetic instructions: i.e. the expression of specific genes in specific regions (or cells) at specific stages. This is most important during pre- and neo-natal brain development. A high degree of flexibility in that development is required throughout life as the brain continues to learn and remember, so functional development of the brain is also likely to be mediated by a variety of environmental factors including maternal and postnatal experiences. Therefore continued brain development is dependent on neural activity (synaptic connection, selection and consolidation, etc.) that allows the brain to become a highly complex organ responsible for memory, learning, consciousness and mind.
New doors have opened in the post-sequencing era onto a better understanding of the design of the brain. As a result, our laboratory deciphered the genetic blueprint for the postnatal development of the mouse cerebellum that can be used as a model to decode the developmental blueprints to other brain regions. Using differential display and microarray techniques, we conducted genome-wide analysis of temporal gene expression patterns and analyzed the spatial (cell-specific) gene expression patterns using in situ hybridization. Finally, we created an integrated database “Cerebellar Development Transcriptome Database (CDT-DB)” (Fig. 1) of the spatio-temporal gene expression profile data. This database will be publicly available this spring. In addition to enhancing the CDT-DB, we are also planning to clarify the gene network involved in each event in development (cell proliferation, synapse formation, etc.). The genetic mechanisms that enable adaptive development in response to environmental variability will be investigated by analyzing the relationships between early life experiences (and epigenetic factors) and gene expressions during the development.
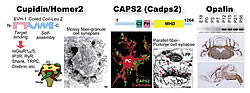
Our laboratory is also investigating the molecular and cellular mechanisms of the brain development. In the course of the CDT-DB project, we discovered a large number of new gene candidates that participate in brain development. We are now exploring the functions of these genes by cloning and allotting several candidate genes to a researcher. The proteins identified up to the present include the Cupidin/Homer2 (postsynaptic scaffolding protein) (Shiraishi et al., 1999, 2003, 2004), CAPS2 (Ca2+-dependent activator protein for secretion 2) (Sadakata et al., 2004), and Opalin (oligodendrocytic paranodal loop protein) (Yoshikawa et al., unpublished) (Fig. 2).
We recently found that CAPS2 is involved in the activity-dependent secretion of neurotrophins, which is indispensable for the differentiation and survival of neurons. Dr. Tetsushi Sadakata discovered that this gene has a peak mRNA expression in granule cells of mouse cerebellum at 7 days after birth. As a homologue CAPS1 is involved in Ca2+-dependent exocytosis of dense-core vesicles that includes catecholamines and peptide hormones, etc., the role of CAPS2-mediated vesicle exocytosis was considered. The neurotrophic factors expressed in granule cells include BDNF (brain- derived neurotrophic factor) and NT-3 (neurotrophin-3), both of which are essential for the cerebellar development, but their secretion mechanisms remain poorly understood. Dr. Sadakata found that CAPS2 is located in the vesicles containing BDNF and NT-3 at the parallel fiber terminals of granule cells connecting Purkinje cell dendritic spines. Using PC12 cells and cerebellar primary cultures, he determined CAPS2’s role in the secretion: it controls activity-dependent secretion of BDNF and NT-3. He also found that, when overexpressed in cultured granule cells, exogenous CAPS2 promoted postsynaptic Purkinje cell survival. These findings indicate that CAPS2 is involved in the differentiation and survival of cerebellar neurons by controlling the activity-dependent secretion of neurotrophins (Sadakata et al., 2004). In addition, Dr. Sadakata is now studying the role of the CAPS2-mediated BDNF secretion in the hippocampus and cerebral cortex where CAPS2 and BDNF are co-localized. Identifying a CAPS2-dependent secretion is first step towards revealing the underlying molecular mechanism of the BDNF and NT-3 secretion. Further analyses with CAPS2 knockout mice should uncover the role of CAPS2-dependent BDNF/NT-3 secretion in the brain development and function.
Our laboratory seeks to elucidate the molecular and cellular mechanisms underlying the brain development through productive, interdisciplinary, and collaborative research projects.